🧬 Introduction: A New Chapter in Life Creation
What if proteins—the complex molecules that drive life—could be created entirely from scratch, not by nature, but by artificial intelligence?
This is no longer a thought experiment. It’s reality.
In a groundbreaking leap for biotechnology and synthetic biology, researchers at EvolutionaryScale have developed esmGFP, the world’s first AI-designed protein that doesn’t exist in nature. Created using their cutting-edge model, ESM3, esmGFP demonstrates that artificial protein design is no longer limited to tweaking existing biological molecules—it can now involve generating entirely new functional proteins through machine learning.
The esmGFP protein, short for evolutionarily scaled model-generated green fluorescent protein, is not just an experimental novelty. It fluoresces like natural GFP, a protein commonly used in cellular imaging, but was created solely by AI, without being modeled after any natural template. This breakthrough represents a paradigm shift: rather than evolving proteins slowly over millions of years, AI can simulate vast evolutionary landscapes and generate new proteins in days or even hours.
Unlike traditional protein engineering, which typically modifies or optimizes natural proteins, esmGFP was created ab initio—from first principles—using ESM3, a large-scale language model trained on hundreds of millions of protein sequences. This process mirrors how ChatGPT generates human-like language, but instead, ESM3 generates biologically viable protein sequences.
This marks the dawn of AI-designed proteins as a new class of biological innovation, with vast implications for medicine, materials science, agriculture, and beyond.
In this blog, we’ll explore:
- What esmGFP is and how it works
- The role of ESM3 in artificial protein design
- Why this development could transform drug discovery, therapeutics, and bioengineering
- And how AI could soon become a designer of life itself
🌟 What Is esmGFP? The First of Its Kind
In the expanding world of synthetic biology, one of the most exciting recent breakthroughs is esmGFP—a green fluorescent protein entirely designed by AI.
Unlike traditional proteins found in nature, esmGFP does not originate from any known organism. It is the product of an AI-designed protein pipeline developed by EvolutionaryScale, using their large-scale transformer model, ESM3. This model was trained on a massive dataset of evolutionary protein sequences, enabling it to predict, generate, and optimize new proteins that mimic biological functions—but were never evolved by nature.
💡 What Makes esmGFP Unique?
esmGFP (Evolutionarily Scaled Model-Green Fluorescent Protein) was created with no input from existing GFP genes. Yet, like natural GFPs—commonly found in jellyfish and widely used in cellular imaging and molecular biology—esmGFP emits bright green fluorescence under ultraviolet light. Its sequence, however, is entirely novel.
🧬 Key Properties of esmGFP:
- Fluorescent behavior comparable to natural GFP
- Stable three-dimensional structure, confirmed by predictive folding tools
- AI-generated amino acid sequence with no direct natural counterpart
- Successful lab synthesis and experimental validation, proving its biological viability
The esmGFP sequence was generated entirely in silico (inside a computer), then tested in vitro, where it folded correctly and fluoresced—just like its naturally evolved cousin. This experiment validated that AI models can move beyond prediction and toward functional protein generation.
🔬 esmGFP vs Natural GFP: A Side-by-Side Comparison
| Feature | esmGFP (AI-Designed) | Natural GFP (e.g., Aequorea victoria) |
|---|---|---|
| Origin | Artificial, designed by ESM3 | Naturally occurring in jellyfish |
| Sequence Similarity | No known homologs | Evolutionarily conserved |
| Fluorescence | Bright green under UV light | Bright green under UV light |
| Structure | Predicted + validated folding | Naturally evolved folding |
| Design Process | Generated via protein language model | Natural selection over millions of years |
🔗 Scientific Validation & Sources
- EvolutionaryScale’s official esmGFP publication
- Preprint on bioRxiv: “Language models enable generative protein design with experimental validation”
- Structure visualizations using AlphaFold & Foldseek comparisons
🤖 Meet ESM3: The AI That Evolved Life Digitally
At the heart of the esmGFP breakthrough lies a cutting-edge AI system called ESM3, developed by EvolutionaryScale. This ESM3 language model isn’t just another protein prediction tool—it’s an entirely new class of transformer-based AI designed to simulate molecular evolution, generate new protein sequences, and predict their structure and function, all without needing a biological blueprint.
🧠 What Is ESM3?
ESM3 (Evolutionary Scale Model 3) is a large language model (LLM) trained specifically on billions of amino acid sequences—the “language” of proteins. Much like ChatGPT understands patterns in human language, ESM3 deciphers and generates protein sequences by learning the grammar of evolution.
But ESM3 doesn’t just mimic existing proteins. It’s capable of molecular evolution simulation—generating brand new sequences that fold into viable 3D structures and even perform biological functions.
🔬 Protein Language Models: ChatGPT for Life Itself
The idea of protein language models is rooted in the same transformer architecture that powers natural language models like ChatGPT. However, instead of being trained on books or websites, ESM3 was trained on massive protein sequence databases such as UniProt and MGnify.
This allows ESM3 to:
- Understand protein syntax and evolutionary patterns
- Predict how sequences fold into 3D structures
- Generate de novo proteins with specific properties (like fluorescence, binding affinity, stability)
It’s a leap beyond structure prediction tools like AlphaFold—because ESM3 doesn’t just predict what proteins look like. It creates them.
🧬 Simulating 500 Million Years of Evolution—In Silico
To design esmGFP, ESM3 effectively condensed half a billion years of natural selection into a few computational steps. It did this by:
- Generating a wide range of sequence variants through deep learning
- Predicting their structures and likely functions
- Selecting sequences with the highest probability of folding into a stable, functional protein
- Sending top candidates for lab synthesis and validation
💡 Think of it as Darwinian evolution—but at machine speed and planetary scale.
🔁 ESM3’s Design Workflow
📌 Diagram:
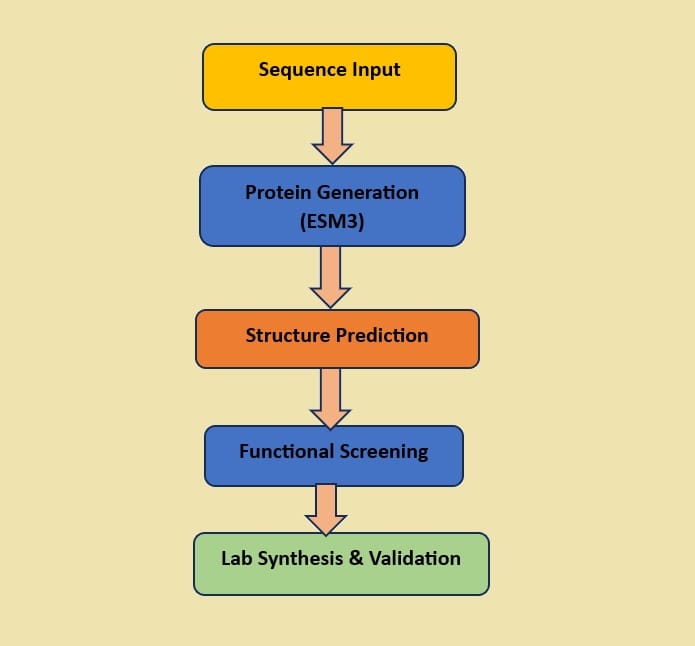
This closed-loop design process allows AI-designed proteins to move from digital concept to real-world biology in weeks instead of millennia.
🌟 Why ESM3 Is a Breakthrough in Synthetic Biology
Unlike traditional bioinformatics tools or even predictive models like AlphaFold, EvolutionaryScale’s ESM3 represents a generative AI system that is capable of creating entirely new biological functions.
It doesn’t just analyze evolution—it evolves digital life.
🔄 How AI Designs Proteins: The Workflow Explained
AI-designed proteins like esmGFP represent a radical shift in how biology is created. Traditionally, designing new proteins involved years of trial-and-error in wet labs—evolving sequences, testing functionality, and iterating through countless mutations. But now, with artificial protein design powered by deep learning, this process can be streamlined, accelerated, and scaled like never before.
At the core of this transformation is a multi-step AI-driven protein engineering pipeline, which blends biological data with computational innovation. Here’s how it works:
🧠 Training: Learning from the Language of Life
The process begins with training a large protein language model—like ESM3—on billions of amino acid sequences collected from across the tree of life. This gives the AI an understanding of:
- How sequences are structured
- Which mutations are viable
- Which motifs are likely to result in functional proteins
Just like ChatGPT learns grammar, ESM3 learns molecular biology.
⚙️ Generation: Designing Novel Sequences
Next, the AI begins generating entirely new protein sequences that don’t exist in nature. These sequences are not random—they are optimized for stability, foldability, and biological plausibility. The goal is to mimic the evolutionary logic of natural proteins while pushing beyond known possibilities.
🧬 Folding Prediction: Simulating 3D Structures
Once a new sequence is proposed, it’s passed through a structure prediction model—often inspired by AlphaFold—to assess whether it can fold into a stable, functional 3D shape. Proteins that fail to fold properly are discarded. This ensures only viable scaffolds move forward.
🔍 Function Simulation: Evaluating Potential Use
Using additional computational models, the AI simulates:
- Fluorescence properties (as in the case of esmGFP)
- Binding affinity to targets
- Enzymatic activity
- Thermal or pH stability
These simulations rank the most promising candidates for lab testing.
🧪 Lab Testing: From Code to Reality
Top-ranked protein designs are synthesized in the lab, expressed in living cells (e.g., E. coli or human cells), and tested for:
- Correct folding
- Fluorescence (in the case of esmGFP)
- Functional behavior
esmGFP successfully passed all key benchmarks: it folded correctly, fluoresced, and demonstrated experimental functionality—despite being entirely AI-generated.
⏱️ The Real Win: Speed, Cost, and Scale
This AI-driven process reduces:
- Time — from years to weeks
- Cost — by avoiding failed wet-lab experiments
- Labor — automating early-stage protein discovery
Traditional wet-lab evolution could take decades to stumble upon a useful protein like GFP. With AI-designed proteins, the same outcome can now be achieved orders of magnitude faster.
This is not just faster science—it’s a fundamentally new way to build life.
🌐 Why esmGFP Matters: Applications and Impact
The development of esmGFP isn’t just a milestone in artificial protein design—it’s a proof of concept that AI in biotechnology can move beyond prediction into the realm of creation. For the first time, we’re seeing synthetic biology evolve into generative biology, where machine learning models like ESM3 don’t just study life—they build it.
But the true value of esmGFP lies in its real-world implications—and how it opens the door to revolutionary applications in medicine, the environment, and agriculture.
💊 Drug Discovery: Faster Enzymes, Better Antibodies
The pharmaceutical industry invests billions of dollars and years into identifying and optimizing proteins that can serve as drugs or drug targets. With AI-powered protein design applications, this process can be:
- Accelerated: AI can instantly propose and simulate thousands of potential drug-protein interactions.
- Optimized: Therapeutic proteins like enzymes and monoclonal antibodies can be redesigned for better specificity, reduced side effects, or longer half-life.
With models like ESM3, researchers could one day generate custom proteins for rare diseases—in weeks, not decades.

💉 Vaccine Development: Building Immune-Targeted Proteins
AI-designed proteins can serve as precise immune system triggers, making them ideal for next-gen vaccine platforms. Unlike traditional vaccines that rely on weakened viruses or fragments of existing pathogens, AI can:
- Create synthetic antigens optimized for stability and immune response.
- Improve vaccine design for mutating viruses (like flu or coronavirus variants).
- Enhance delivery efficiency with custom protein carriers.
esmGFP proves that non-natural proteins can be made stable, functional, and biocompatible—core requirements for future synthetic vaccines.
🌱 Environmental Biotech: Cleaning Up with Code
One of the most exciting areas of protein design applications is environmental biotechnology. Enzymes created through AI could:
- Break down plastic waste faster and more efficiently
- Capture and sequester CO₂ from industrial exhaust
- Detoxify contaminated soil or water with tailored bio-remediation proteins
With models like ESM3, we could one day program enzymes to clean oceans, purify polluted rivers, or restore damaged ecosystems—using proteins that never existed before.
🌾 Agriculture: Engineering Proteins for Crop Resilience
In agriculture, AI-designed proteins can help create:
- Custom plant proteins that improve drought tolerance or pest resistance
- Biofertilizers and growth enhancers that reduce the need for synthetic chemicals
- Enzymes that break down soil toxins or improve nutrient absorption
As climate challenges grow, the ability to custom-design proteins for specific agricultural conditions could transform food security worldwide.
🧬 esmGFP: The Proof AI Can Build Biology
Before esmGFP, protein language models were mostly used to analyze or predict biology. esmGFP changes that. It demonstrates experimentally that AI can:
- Generate novel protein sequences
- Predict their structure
- Simulate their function
- And synthesize them successfully in the lab
🧠 “esmGFP is the first real evidence that generative AI can produce functional biological molecules,” said the team at EvolutionaryScale, in their public release on esm3.
💬 In a viral post on X (Twitter), one researcher commented: “This is the ChatGPT moment for biology—except instead of writing essays, it’s writing life.”
📣 Call to Action:
Imagine a world where AI creates enzymes to clean oceans or cure rare diseases. That world just got a little closer with esmGFP.
🚀 Generative Biology: A New Frontier
We’ve entered the era of generative biology—a groundbreaking field where life itself can be designed by algorithms.
Much like how AI can now create essays, artwork, or even music, generative biology uses large language models and deep learning to create new biological structures, such as proteins, enzymes, or potentially entire organisms. The key difference? Instead of generating pixels or paragraphs, this AI builds with amino acids, folding patterns, and functional properties.
🧬 What Is Generative Biology?
Generative biology refers to the use of AI to generate new biological molecules, especially proteins, that do not exist in nature. These aren’t mere predictions—they are algorithmically created instructions for functional life components.
This concept is made possible by training AI models on vast databases of evolutionary and molecular data, allowing them to understand:
- The “grammar” of amino acids
- The “syntax” of protein structures
- And the “meaning” of biological function
The result? AI-designed proteins like esmGFP, capable of folding, fluorescing, and functioning despite never having evolved in nature.
🤖 Parallels to Other Generative Fields
To understand how disruptive this is, consider how AI has already transformed:
- Language: GPT models generate human-like text
- Art: DALL·E and Midjourney create original images from prompts
- Music: AI composes full symphonies
- Video: Sora and Runway generate lifelike visual sequences
Generative biology is simply applying that same deep learning power to molecular biology—with much higher stakes.
🧠 ESM3 vs ESMFold vs AlphaFold: Who’s Leading?
Several major AI players are now racing to lead the next frontier of biology:
| Model | Developer | Focus | Strength |
|---|---|---|---|
| ESM3 | EvolutionaryScale | Generative protein design | Creates and validates novel proteins |
| ESMFold | Meta AI | Protein structure prediction | Fast, scalable folding model |
| AlphaFold 2 | DeepMind (Google) | Accurate structure prediction | Best-in-class structural accuracy |
- AlphaFold revolutionized structural biology by predicting how proteins fold.
- Meta’s ESMFold made structure prediction even faster, suitable for large-scale scans.
- But EvolutionaryScale’s ESM3 goes a step further—it doesn’t just predict, it creates.
That’s why esmGFP isn’t just a novelty—it’s a proof that AI can build functioning biology, not just analyze it.
🔮 What’s Next in Generative Biology?
The future potential of AI in molecular biology is staggering:
- Programmable proteins tailored for specific roles in medicine, industry, or the environment
- Custom-designed enzymes for breaking down pollutants, processing food, or building materials
- Next-gen therapeutics built from scratch for hard-to-treat diseases
- AI-driven evolution, where instead of waiting millions of years, we design new organisms in weeks
This intersection of AI + biology will likely define the next decade of biotech innovation.
Generative biology is not just a tool—it’s a new language for designing life.
⚖️ Ethical and Safety Considerations
As the power of AI-designed proteins expands, so too does the responsibility to ensure that we understand and control it. The rise of generative biology isn’t just a scientific triumph—it’s an ethical frontier.
🧬 Could AI Create Harmful or Uncontrollable Proteins?
One of the most pressing concerns in bioethics for AI proteins is dual-use risk. While AI can design proteins for medicine and sustainability, the same tools could be misused to:
- Generate toxic proteins, pathogens, or bioweapons
- Create unregulated synthetic organisms that escape containment
- Disrupt ecosystems by introducing proteins with unintended effects on life systems
Because models like ESM3 can operate with minimal biological input, there’s concern about bad actors using open-source tools for unethical applications.
🧬 Who Owns AI-Generated Biology?
Another major debate centers on intellectual property:
- Can a protein invented by an algorithm be patented?
- Does ownership belong to the developers, the data providers, or the model itself?
- Should life created by AI be open science or commercially locked down?
Some argue that AI-generated proteins should follow the norms of traditional biotech patents. Others believe they should be open-access breakthroughs for the good of humanity—especially when designed using public datasets.
🛑 Should Generative Biotech Be Regulated?
The field of synthetic biology already faces tight regulations, especially in pharma and agriculture. But AI-driven molecular design introduces a layer of complexity regulators are still grappling with:
- Do we need global oversight similar to what’s being proposed for AI in general?
- How do we validate and monitor AI-designed proteins once released into real-world systems?
- What frameworks will ensure transparency, traceability, and accountability?
A global regulatory dialogue—involving governments, scientists, ethicists, and AI companies—is urgently needed to guide the responsible use of these technologies.
🌐 Open Science vs Proprietary AI in Biology
The open release of AlphaFold and ESMFold helped democratize access to structural biology. However, commercialized models (like ESM3) signal a shift toward private biotech IP generation.
The balance between open science and proprietary control will shape how equitable and ethical this new era of synthetic biology becomes.
⚖️ Generative biology offers incredible benefits—but must be tempered with foresight, regulation, and responsibility.
🧬 Conclusion: Have We Witnessed the Birth of AI Life?
The creation of esmGFP by EvolutionaryScale isn’t just another scientific milestone—it may be one of the most profound events in the history of biotechnology.
For the first time, an AI system didn’t just analyze a protein or predict its structure—it created one from scratch, guided only by patterns learned from evolution. The result? A functional, fluorescing, experimentally validated protein—an artificial molecule that never existed in nature.
This wasn’t an optimized tweak or a lab-modified variant. esmGFP was a pure product of AI-driven generative biology, built with no natural blueprint, and validated through both computation and experiment.
🌍 A Paradigm Shift in Protein Design
We’re now witnessing the convergence of synthetic biology and artificial intelligence, where:
- Drug discovery accelerates
- Custom molecules are engineered on demand
- Evolution becomes a computational process, not a natural one
What once took millennia of random mutations can now be designed in weeks by algorithms.
🌱 What esmGFP Means for the Future
This single protein—built entirely in silico, then brought to life in a lab—marks a pivotal moment:
- It validates that AI-designed proteins can fold, function, and fluoresce
- It shows that intelligent design of biology is no longer metaphorical—it’s literal
- It suggests a future where digital life engineering becomes routine in medicine, sustainability, and beyond
esmGFP is not just a protein—it may be a glimpse into the future of life engineered by intelligence.
🧬 Curious about more biotech breakthroughs powered by science and innovation?
Explore these cutting-edge advances shaping the future of medicine:
-
💉 Promising Results of the mRNA Cancer Vaccine in Trials – Discover how personalized mRNA therapies are already delivering hope in human cancer studies.
-
💊 Methylene Blue: Bridging Medicine and Future Technology – A 19th-century compound making a 21st-century comeback in neuroprotection and longevity research.
-
⚛️ Terbium-161: Advancing Nuclear Cancer Therapy – See how a next-gen radiopharmaceutical is redefining precision cancer treatment.
Click any title to explore how technology and biology are merging to fight disease like never before.