🧬 Hooked by a Barcode: Introduction to Unique Molecular Identifiers (UMIs)
What if a single typo in your DNA could be the difference between life and death—yet still go unnoticed by even the best sequencing machines? That’s exactly the kind of problem Unique Molecular Identifiers (UMIs) were designed to solve.
Unique Molecular Identifiers (UMIs), often referred to as molecular barcodes, are short, randomized nucleotide sequences that are linked to each individual DNA or RNA strand prior to amplification, enabling precise tracking and quantification. These small tags allow scientists to trace each molecule uniquely, separating real biological variation from PCR duplicates and sequencing errors. In simpler terms, they let us count what’s actually there, not what appears to be.
This matters—especially in ultra-sensitive medical diagnostics. In cancer genomics, for instance, UMIs have enabled early detection of ultra-rare mutations, helping identify diseases long before symptoms appear. In one real-world case, researchers used UMIs during liquid biopsy RNA-seq to spot a mutation present in fewer than 0.01% of all sampled DNA—a signal that would have otherwise been drowned in noise.
But this is just the beginning. As UMIs become standard in next-generation sequencing (NGS), they are opening the door to radically more accurate diagnostics, drug development, and personalized medicine. And in the near future, the very idea of health monitoring could shift—from routine blood tests to barcode-based molecular snapshots of your biology, captured in real time.
In this post, we’ll break down what UMIs are, how they work, where they’re already saving lives, and where they’re heading next.
🧪 What Are molecular barcodes in sequencing and Why They Matter
In the era of next-generation sequencing (NGS), precision is everything. Whether you’re detecting rare cancer mutations, studying gene expression in single cells, or analyzing complex microbial populations, noise from amplification errors and PCR bias can easily skew your results. This is where Unique Molecular Identifiers (UMIs)—also known as molecular barcodes in sequencing—come into play.
So, what is the UMIs definition in technical terms? A UMI is a short, randomly generated sequence of nucleotides (usually 8–12 bases) that is attached to individual DNA or RNA molecules during the early stages of sequencing library preparation—before PCR amplification. This small tag functions like a molecular fingerprint, uniquely identifying the original source molecule from which the read originated.
Here’s why this is revolutionary:
- Accurate PCR duplicate removal: Without UMIs, you can’t distinguish between a genuine highly-expressed gene and multiple PCR-amplified copies of the same transcript. UMIs allow you to collapse all duplicate reads (those with identical mapping coordinates and UMI tags) into a single “original” molecule, providing a clearer, undistorted signal.
- Improved quantification: In applications like RNA-seq or ctDNA analysis, correct molecule counting is crucial. UMIs help ensure that your quantification reflects biology, not PCR artifacts.
- Error correction: By comparing sequences from reads sharing the same UMI, sequencing errors can be corrected, improving confidence in variant calling—especially important in detecting ultra-rare variants in low-frequency populations, such as early cancer markers in liquid biopsies.
- Single-cell sequencing advantage: In single-cell studies, every molecule counts. UMIs help track the transcriptomic fingerprint of individual cells, revealing subtle differences that would be masked without accurate molecular labeling.
In summary, molecular barcodes in sequencing, specifically UMIs, aren’t just another feature—they’re becoming essential. From PCR duplicate removal to robust single-cell resolution, UMIs are shaping the future of genomics by making every molecule matter.
🔬 How UMIs Improve Accuracy in NGS Workflows
In modern next-generation sequencing (NGS), speed and scale are no longer the bottleneck—accuracy is. Researchers and clinicians now seek results they can trust down to the rarest molecule. But traditional workflows introduce critical flaws—mainly PCR amplification errors and bias. Enter Unique Molecular Identifiers (UMIs), which are redefining NGS error correction by offering molecule-level precision.
🔁 Amplification Bias and PCR Errors
When DNA or RNA samples are prepared for sequencing, they undergo PCR amplification to generate enough material for detection. Unfortunately, this introduces two main issues:
- Overcounting: The same molecule may be amplified hundreds of times. Without a way to trace it back to its origin, the data overestimates abundance.
- PCR amplification errors: Mutations can be accidentally introduced during copying, making them indistinguishable from true genetic variants.
These flaws make accurate quantification—and especially detection of low-frequency variants—nearly impossible without correction.
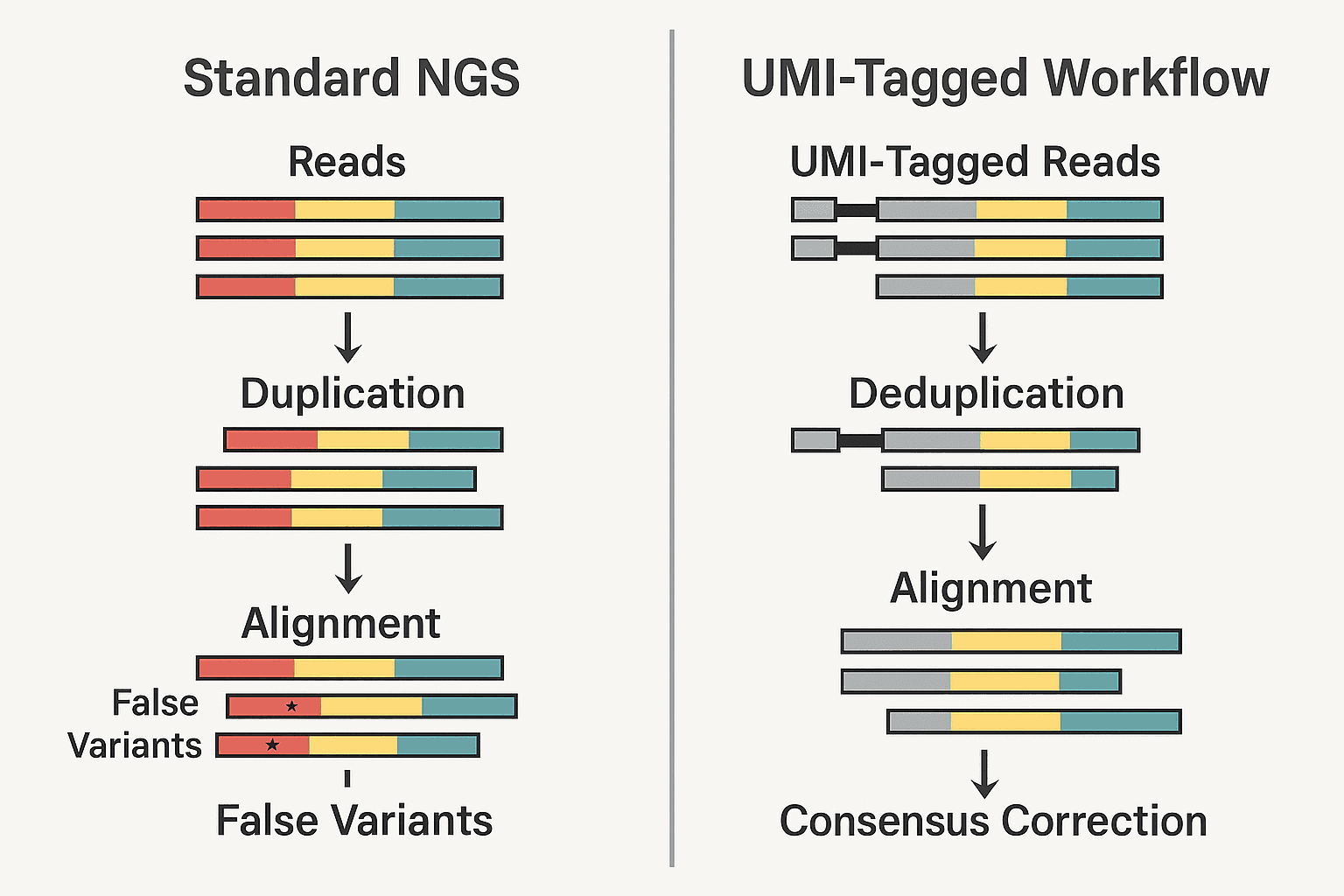
🎯 NGS Error Correction with UMIs
UMIs offer a powerful fix. Because each molecule is tagged with a unique sequence before amplification, reads derived from the same original molecule can be grouped together:
- Deduplication: By collapsing all identical UMI-labeled reads at the same genomic location into a single representative, researchers eliminate inflated counts.
- Consensus calling involves aligning all reads that share the same UMI to detect and correct sequencing or PCR-induced errors, ensuring more accurate and reliable data. This provides a corrected “consensus” sequence that more accurately reflects the truth.
This approach transforms noisy data into clean, trustworthy insights—especially when sequencing degraded, low-input, or rare samples.
🔍 Low-Frequency Variant Detection
One of the most transformative uses of UMIs is their ability to detect rare genetic variants with high accuracy, even when present at extremely low frequencies. In cancer genomics and liquid biopsies, a handful of mutated DNA fragments may be hiding among millions of healthy ones. Similarly, in rare disease research, accurate variant calling from limited DNA is critical.
By using NGS error correction through UMIs, researchers can reliably detect variants occurring at frequencies as low as 0.1%—a feat impossible with traditional pipelines. UMIs also increase sensitivity in circulating tumor DNA (ctDNA) analysis, MRD monitoring, and non-invasive prenatal testing (NIPT).
🧫 Real-World Applications of UMIs in Genomics
Unique Molecular Identifiers (UMIs) are no longer just theoretical tools—they are being deployed across a growing number of genomic fields, unlocking breakthroughs in disease detection, drug development, and precision medicine. These UMI use cases show how powerful molecular barcoding has become in real-world settings:
🧠 Single-Cell RNA sequencing UMIs: Precision at the Cellular Level
In single-cell RNA sequencing UMIs, where individual cells are profiled to study gene expression, noise and amplification bias are rampant. Without UMIs, it’s nearly impossible to distinguish between a gene truly expressed multiple times or simply overamplified by PCR.
- UMIs tag each RNA molecule before amplification, enabling true quantification.
- This is critical for identifying rare cell types, understanding developmental biology, and tracking gene expression in diseases like cancer or neurodegeneration.
The use of single-cell RNA sequencing UMIs is now standard in platforms like 10x Genomics and Smart-seq3, allowing accurate cell-level resolution across complex tissues.
🧬 Liquid Biopsy and Cell-Free DNA in Cancer
In liquid biopsies, minuscule fragments of tumor-derived DNA—known as circulating tumor DNA (ctDNA)—float freely in the bloodstream, offering a non-invasive window into cancer’s genetic makeup. Detecting these fragments—especially at early stages—requires ultra-sensitive tools.
- UMI in liquid biopsy allows researchers to distinguish true low-frequency mutations from sequencing noise.
- Used in monitoring minimal residual disease (MRD), cancer relapse, and treatment resistance.
For example, Guardant Health and Foundation Medicine use UMI-enhanced workflows to enable non-invasive cancer diagnostics with unprecedented precision.
🦠 Viral Diversity and Antiviral Resistance
Viruses like HIV and SARS-CoV-2 mutate rapidly, and understanding that diversity is crucial for vaccine and drug development.
- UMIs help track individual viral genomes, removing duplicate sequences and filtering out artificial variants caused by PCR errors.
- Researchers can now map viral quasispecies—genetically distinct variants within a single host—more accurately than ever.
This has been especially useful in COVID-19 variant surveillance and HIV resistance profiling.
🧪 CRISPR and Gene Editing Validation
One of the lesser-known but rapidly growing UMI use cases is in CRISPR-Cas9 validation.
- After gene editing, UMIs ensure that detected changes are true edits, not errors introduced during amplification or sequencing.
- They improve precision in assessing off-target effects—vital for therapeutic genome editing.
Biotech labs and startups now routinely use UMIs for CRISPR QC pipelines, especially in clinical-grade applications.
By enabling molecular-level accuracy, UMIs are pushing genomics into a future where data is not just big—but deeply trustworthy.
🚲 The Future of UMIs – Cellular Barcodes for Living Systems
What began as a tool for sequencing accuracy may soon become the blueprint for living, self-documenting systems. As synthetic biology, neurogenomics, and nanotech converge, the future of Unique Molecular Identifiers (UMIs) could evolve far beyond today’s PCR deduplication workflows. Here’s where science meets visionary engineering:
🧬 Live UMIs: Self-Generated Barcodes in Living Cells
Imagine engineered cells that generate molecular barcodes on their own, tagging internal biological events—like stress responses, infections, or mutations—in real time.
- These “live UMIs” could serve as molecular black boxes, allowing scientists to reconstruct a cell’s history.
- CRISPR-based “recording systems” like mSCRIBE and DNA Tape Recorders at Harvard and MIT are already heading in this direction.
- In the future, these tags might help diagnose early cancer development before symptoms appear—just by decoding a cell’s barcode log.
🧠 Brain UMI-Mapping: Logging Thought in Molecular Code
The brain is the most data-rich organ in the body, and yet we have no molecular way to track its synaptic activity at scale. Now imagine:
- UMIs engineered into RNA that tag neural firing patterns at specific synapses.
- These tags could create a permanent record of activity—useful for mapping memory formation, learning, or mental illness biomarkers.
- DARPA-funded projects like Neurogenetic Recording hint at this possibility, where each synaptic event is cataloged like a digital journal.
🤖 DNA-Based Digital Memory: Biological USB Drives
As the line blurs between biology and computing, DNA is emerging as the next data storage medium—and UMIs could make it dynamic.
- Think artificial DNA circuits where UMIs act as write-once, read-many memory units, storing environmental data or system states.
- Future robots or biosensors could use DNA memory storage to track exposure to toxins, radiation, or other conditions over years.
Microsoft and Twist Bioscience are already exploring petabyte-scale DNA storage—with molecular barcodes enhancing retrieval and reliability.
👩⚕️ Self-Updating Health Records: Implanted UMIs
In tomorrow’s hospitals, your medical file may not live in a cloud—but inside your body.
- UMIs could be implanted as biosafe patches or embedded in circulating cells, continuously tracking genetic mutations, immune shifts, or viral loads.
- Over time, these molecular barcodes would build a self-evolving health record, alerting you to changes before symptoms arise.
- Integration with wearable devices and AI could turn this into real-time molecular tracking, radically transforming personalized medicine.
The future of UMIs lies not just in reading life’s code—but in helping it write its own story, molecule by molecule.
🧱 Challenges and Limitations of UMI Technology
While Unique Molecular Identifiers (UMIs) have revolutionized next-generation sequencing (NGS) accuracy, the technology still faces several critical challenges that must be addressed before it sees wider clinical adoption.
🔄 Sequence Collisions and Short UMI Limitations
One of the most pressing UMI sequencing problems is collision—when two different molecules randomly receive the same barcode. This is more likely when using short UMIs (e.g., 4–8 nucleotides), which have a limited number of combinations. Collisions compromise deduplication accuracy, particularly in high-throughput datasets like single-cell RNA-seq or liquid biopsy, where molecule diversity is extremely high.
🧮 Pipeline Complexity and Data Overhead
Incorporating UMIs introduces significant computational complexity. Traditional pipelines must be adapted to:
- Group reads by UMI,
- Perform consensus calling to correct sequencing/PCR errors,
- And track these across multiple samples.
This adds processing time, memory load, and alignment challenges, especially when working with large datasets or low-frequency variants.
⚙️ Lack of Platform Standardization
Another hurdle is the lack of standardized protocols for UMI integration across platforms like Illumina, Oxford Nanopore, or PacBio. Differences in structured UMIs (barcodes with known sequences vs random ones), adapter designs, and read orientation create inconsistencies that affect downstream reproducibility and interoperability.
While challenges in Unique Molecular Identifiers (UMI) deduplication persist—such as barcode collisions and amplification bias—ongoing advances in UMI-aware algorithms, expanded barcode diversity, and synthetic spike-in controls are steadily enhancing both accuracy and scalability. Still, for UMI-based sequencing to reach widespread clinical adoption, it must first clear these critical technical hurdles.
✅ Conclusion: A Barcode That’s Reshaping Biology
Unique Molecular Identifiers (UMIs) are no longer just a technical upgrade in sequencing—they’re becoming a cornerstone of precision medicine. By tagging individual molecules and filtering out noise, UMIs offer unmatched clarity in detecting mutations, tracking cellular behavior, and validating gene edits. From liquid biopsies to single-cell analysis, UMIs are redefining what’s possible in genomic science.
But this is only the beginning.
As we look to the future, the potential of molecular barcoding for life sciences grows even more powerful. Imagine a world where every cell carries its own digital ID, logging every mutation, stress signal, or therapeutic response in real-time. Could UMIs one day serve as the backbone of a living health record?
If biology is the code of life, UMIs might just be its most important debug tool.
Explore more cutting-edge tech and futuristic science: